Navigation auf uzh.ch
Navigation auf uzh.ch
Our institute owns and operates next generation sequencing (NGS) platforms, which are instrumental to perform high throughput DNA sequencing in human samples. We make use of NGS to detect disease-causing mutations in patients and families with monogenic (Mendelian) diseases. These include eye diseases, cardiomyopathies, long QT syndrome, polyneuropathies, familial fever syndromes and others (genetic testing). All of them are characterized by a significant genetic heterogeneity and single gene sequencing is inefficient for comprehensive genetic analysis.
Retinal and Vitreoretinal Diseases
These disorders are characterized by a tremendous genetic heterogeneity and clinical variability. Currently, mutations in more than 300 genes are associated with about 20 retinal and vitreoretinal diseases (for review please refer to: Berger W, Kloeckener-Gruissem B, Neidhardt J, 2010: The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res 29:335-375; overview). Because of the large number of genes to be screened for mutations, high throughput technologies are required to provide reliable diagnostic testing. The following figures show examples for genetic heterogeneity and clinical variability.
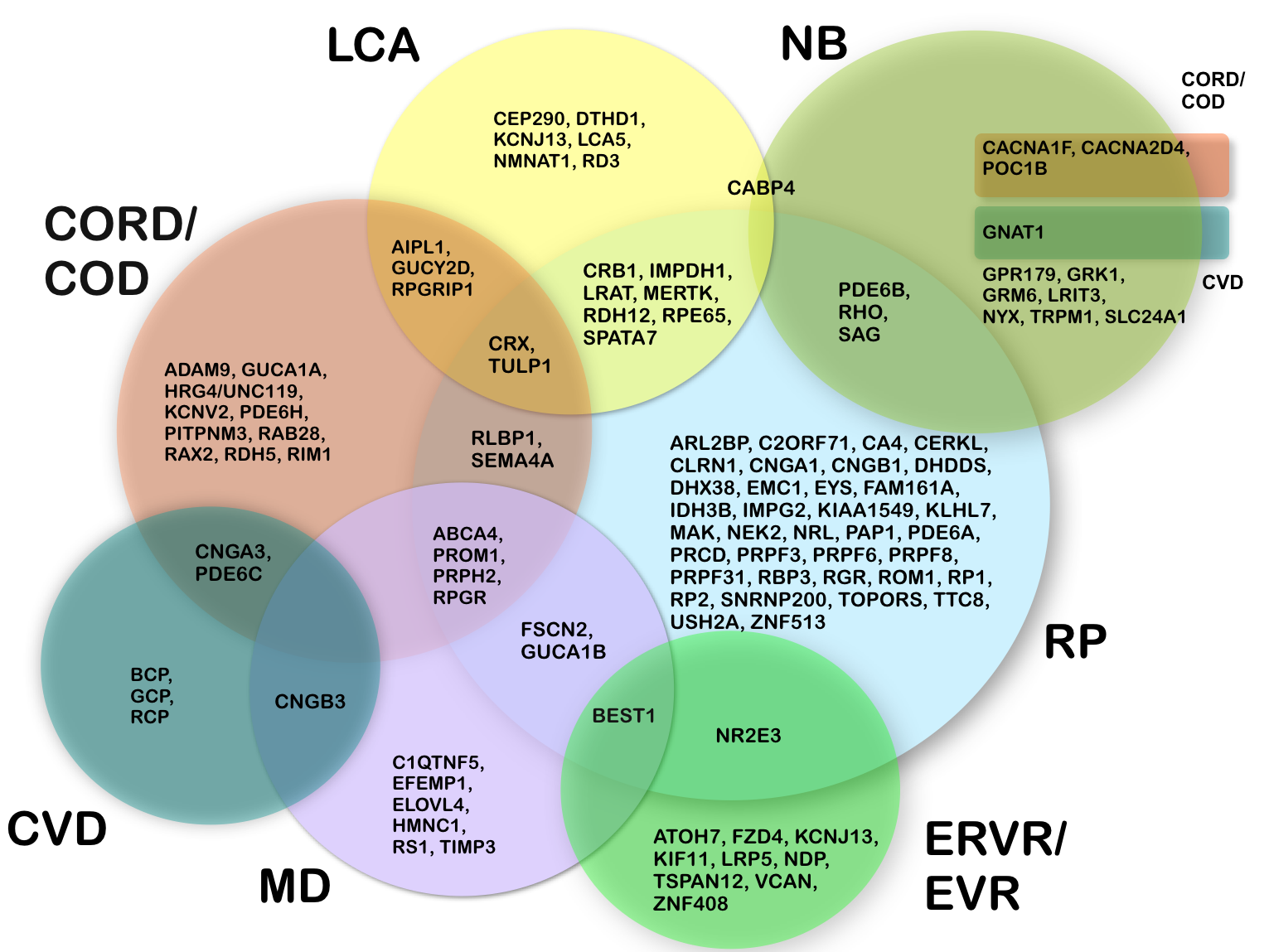
Figure: Clinical variability in monogenic retinal and vitreoretinal diseases. The clinical manifestations of mutations in a single gene can be highly variable. The circles represent different clinical diagnoses. Gene symbols in the overlapping areas indicate that mutations may lead to either of the phenotypes, as for example mutations in ABCA4, which can give rise to RP, MD, COD or CORD.
CORD/COD: cone rod dystrophy / cone dystrophy; CVD: colour vision defect; ERVR/EVR: erosive and exudative vitreoretinopathy; LCA: Leber congenital amaurosis; MD: macular degeneration; NB: night blindness; RP: retinitis pigmentos
In addition to highly variable clinical manifestations of mutations in several genes, the genetic heterogeneity of retinal and vitreoretinal diseases is particularly challenging for genetic testing of patients and families. One of the most prominent examples for this genetic heterogeneity is retinitis pigmentosa (RP). The figure below shows the large variety of genes involved in RP and other diseases, which predominantly involve the rod photoreceptors.
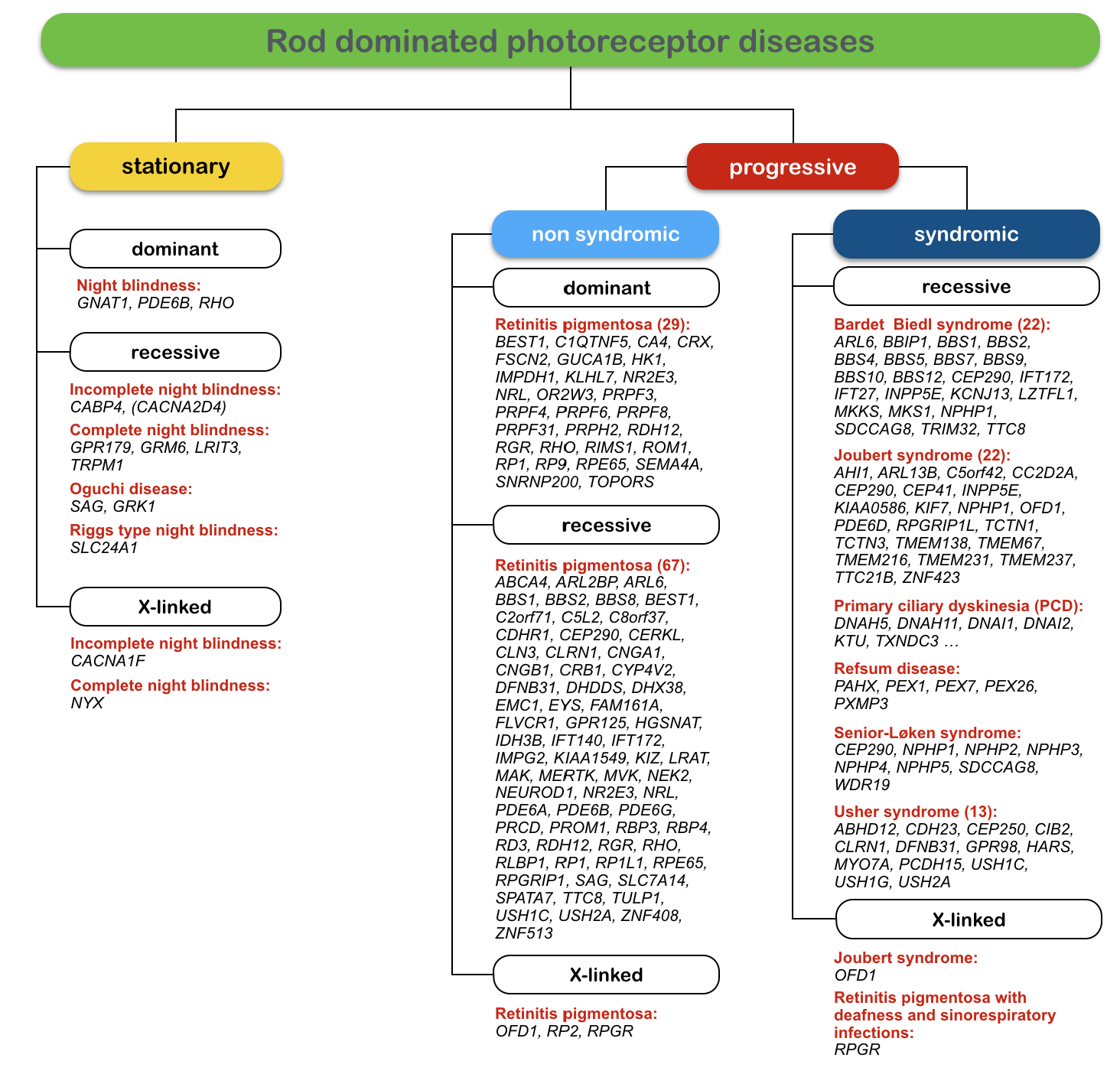
Figure: Genetic heterogeneity of monogenic rod dominated retinal diseases. Rod dominated diseases can be stationary (night blindness) or progressive (retinitis pigmentosa). In syndromic forms, the retinal manifestations are combined with other clinical symptoms as for example deafness in Usher syndrome. The numbers of genes known today (as of September 2015) are given for dominant, recessive and X-linked forms of the diseases. In recessive RP for example, 67 genes were shown to carry disease-causing mutations.